Automatic anatomical analysis of MRI
A comprehensive pipeline inspired by morphology, pix2pix GAN, and SVM
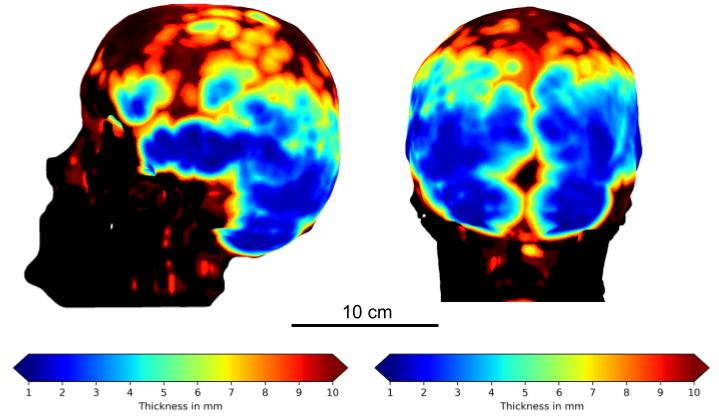
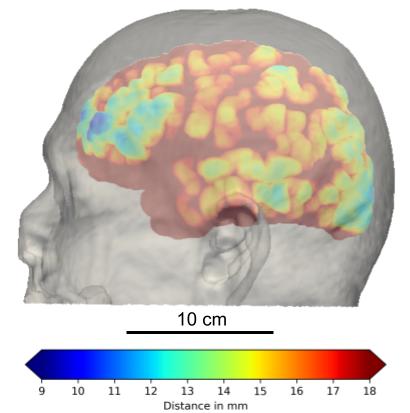
At the request of doctors, I measure the skull thicknesses and scalp-to-cortex distances at four different locations (sphenoid, parietal, etc.) from MRI scans. The data is further categorized in terms of gender and age to study the distribution. This geometric information is important to transcranial imaging modalities involved with optics and ultrasound because they have limited depth and resolution for functional imaging.
Segmentation of skull and calculating its thickness profile from T1 MRI are generally very challenging because bones are black along with air in the scans. The first solution is using morphological transformations to extract the surface of inner and outer surfaces of the skull and calculating the 95% Hausdorff distance of the outer surface. The same procedure is applied to the scalp and cortex. By registering each MRI to a template in advance, we can make sure that we are measuring the same location for each sample. And thanks to the API of 3D Slicer, we can process a batch of scans automatically.
Another solution, which is getting interesting, is using machine/deep learning to segment the skull from MRI. In this project, I calculate a feature-descripting vector (descriptor) for each voxel (3D pixel) of the images based on the (pre-registered) location and neighbor intensities. By training present paired MRI-CT dataset with support vector machine, we optimize the weights that classify each voxel as skull or non-skull. This method works well with limited number of training data like paired MRI-CT. Another possible solution is directly using 3D U-net to segment the skull, which needs to be investigated further.
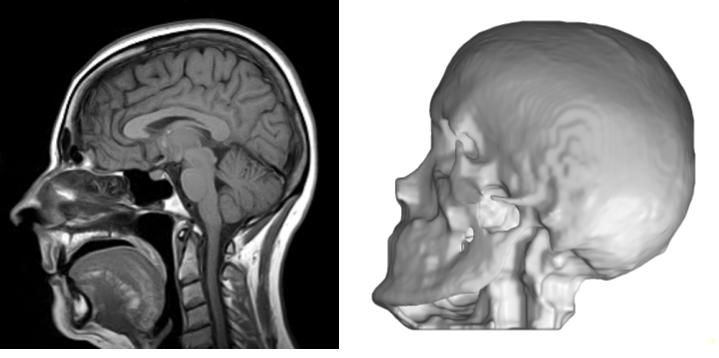
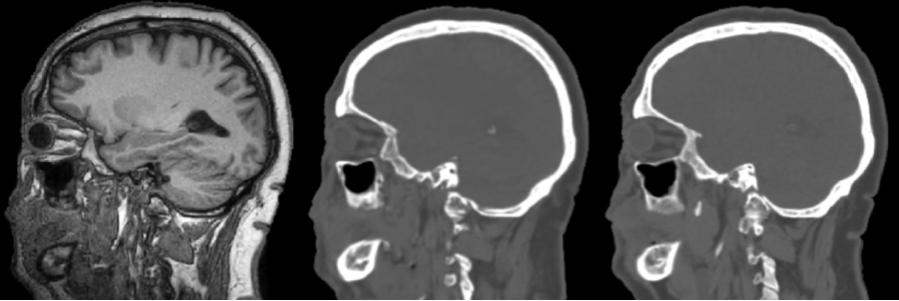
Even more interestingly, I am currently trying to construct a generative adversarial network (GAN) inspired by pix2pix GAN to directly translate 3D MRI to CT, and then segment the skull by plain thresholding. A test result is shown below, but the result is a bit unstable. Perhaps I need to keep tuning the hyperparameters, so stay updated!